Self-organization and positioning of bacterial protein clusters
Many cellular processes require proteins to be precisely positioned within the cell. This is the case even when the proteins are dynamic and cannot rely on existing cellular landmarks. Scientists at the Max Planck Institute for Terrestrial Microbiology in Marburg have proposed a novel physical model for how such dynamic proteins can self-organise and position themselves into regular repeating patterns inside bacterial cells. Their results appeared today in Nature Physics.
While many proteins can be captured by an existing landmark or by sensing membrane curvature, this is not always the case. In rod-shaped bacteria, several protein complexes are regularly positioned along the cell without any known recruitment factor. Seán Murray and Victor Sourjik have proposed a model for how the self-organization and positioning of dynamic nucleoid-associated complexes could be achieved. In short cells the mechanism reliably results in a single focus at mid-cell, while in longer cells a focus is positioned at each quarter-position and so on with increasing length. Furthermore, nucleoid growth leads naturally to foci splitting and dynamic adjustment of the pattern.
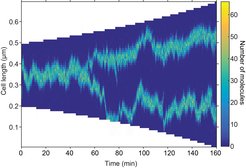
Self-organisation and pattern formation in biology have most often been explained by the Turing mechanism, a physical effect whereby interacting molecules can overcome the homogenizing effect of diffusion to form regions of high density. However, while the effect can explain why a pattern forms, it cannot specify which pattern forms. The authors overcame this problem by combining a Turing mechanism with a positioning mechanism based on exchange of molecules between the chromosome (nucleoid) and the cytosol. This results in the occurrence of only regularly positioned patterns. In essence, the mechanism allows proteins to sense the nucleoid length and position themselves appropriately.
The authors use the model to explain the observed self-organisation of Escherichia coli SMC, an essential protein involved in DNA organisation that is found in all domains of life. However, they suggest that the model’s generality means that it is likely applicable to other regularly positioned, yet dynamic, proteins, such as those regulating cell division in rod-shaped bacteria.
Reference:
Murray, S.M. & Sourjik, V. (2017) Self-organization and positioning of bacterial protein clusters. Nature Physics, 1745-2481












