Complex Adaptive Traits (CATs)
Dr. Ilka Bischofs-Pfeifer
Research area
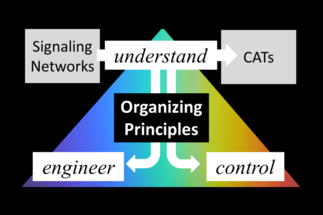
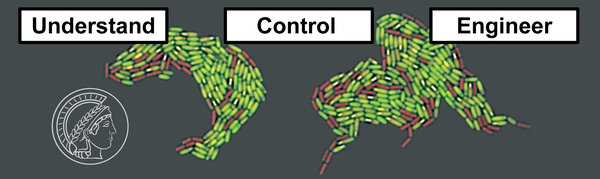
Bacteria have evolved sophisticated strategies to cope with stress and adapt to environmental change. Our group studies complex adaptive traits (CATs) in beneficial Bacilli at the behavioral and mechanistic level. Our research reveals new traits and aims to understand how the gene regulatory network controls them. We specifically focus on spores and peptide-based signaling networks and seek to identify fundamental organizational principles relating the molecular network design to population-level behavior and vice versa. This should facilitate rational manipulations of bacterial populations and the development of probiotics in the future. In cross-disciplinary studies with together with others we also study stress-associated phenomena with relevance for humans and the environment (e.g. the formation of methane) using B. subtilis as a model organism.

Please visit also our website at the BioQuant of the University of Heidelberg.
Selected recent publications
A. Mutlu, C. Kaspar, N. Becker and I. B. Bischofs (2020)
A spore quality-quantity tradeoff favors diverse sporulation strategies in B. subtilis . The ISME J. doi: 10.1038/s41396-020-0721-4
H. Babel, P. Naranjo-Meneses, S. Trauth, S. Schulmeister, G. Malengo, V. Sourjik and I. B. Bischofs (2020)
Ratiometric population sensing by a pump-probe signaling system in Bacillus subtilis. Nature Communications 11, 1176
I. Lytvynenko, H. Paternoga, A. Thrun, A. Balke, T.A. Müller, C.H. Chiang, K. Nagler, G. Tsapraillis, S. Anders, I. Bischofs, J.A. Maupin-Furlow, C. M. T. Spahn and C. A. P. Joazeiro (2019)
Alanine tails signal proteolysis in bacterial ribosome-associated quality control. Cell, 178(1),76-90
A. Mutlu, S. Trauth, M. Ziesack, K. Nagler, J.-P. Bergeest, K. Rohr, N. Becker, T. Höfer and I.B. Bischofs (2018)
Phenotypic memory in Bacillus subtilis links dormancy entry and exit by a spore quantity-quality tradeoff. Nature Communication 6: 69