Hot news from submarine volcanoes: Surprising modifications identified in methane-generating enzymes
The research group of Seigo Shima at the Max Planck Institute for Terrestrial Microbiology in Marburg, together with scientists at the Max Planck Institute of Biophysics in Frankfurt, have a longstanding interest in understanding how methanogens generate methane. They have previously solved the 3D structure of several MCRs and uncovered the trick they use for the methane generation. The key of the reaction is performed by a unique nickel cofactor called F430. Moreover, MCR contains six uncommon amino acids that are modified post-translationaly and might play a role in catalysis.
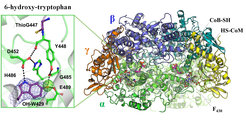
Modified residues in the vicinity of the active site. The post-translationally modified residues 6-hydroxy-tryptophan and thiolglycine are colored in purple and the residues interacting with them are shown in balls and sticks. Hydrogen bonds network (in black dash) connects the 6-hydroxy-tryptophane to the thioglycine (considered as part of the catalysis) backbone. The electron density in blue mesh and the positive density in green mesh are the experimental evidence of the 6-hydroxy-tryptophane presence.
Structural organization of the methane generating enzyme called the methyl-coenzyme M reductase (MCR). MCR contained three subunits (alpha,beta,gamma) outlined by different colors which make dimers (in cyan, yellow and light green). One of the subunit is shown in transparent to highlight the actors of the reaction: coenzyme M, coenzyme B and the cofactor F430 in balls and sticks.
To explore the diversity of MCR and to determine if all methanogens use the same trick, Seigo Shima together with Tristan Wagner, a postdoctoral fellow, determined the 3D structure of the MCR enzymes from two methanogens from submarine volcanoes, Methanotorris formicicus (that lives at 80°C) and Methanothermococcus thermolithotrophicus (that lives at 65°C). These hyper-thermophilic methanogens grow very fast with doubling-times of only 30 min.
The two MCRs from the hyper-thermophilic methanogens are almost identical, and only have a few changes at the protein surface compared to other MCRs. The catalytic site where methane is formed is identical to that in other MCRs, reflecting the chemically challenging mechanism of methane formation. However, a big surprise was waiting for the scientists in the surroundings of the catalytic site of MCR from Methanotorris formicicus. Here, the scientists identified the amino acid tryptophan with an unusual modification. The mass of this tryptophan residue was increased by 16 Da corresponding to an extra oxygen molecule. This 6-hydroxy-tryptophan, has, so far, only been found in deadly toxins from mushrooms. This special tryptophan interacts with parts of the MCR protein that are important for catalysis. Seigo Shima and Tristan Wagner speculate that this unusual modification helps the protein to become more stable and might boost its catalytic power to allow the microbes to survive in the harsh hot marine environment.
For more information
J Bacteriol. 2017 May 30. pii: JB.00197-17. doi: 10.1128/JB.00197-17. [Epub ahead of print]
Phylogenetic and structural comparisons of the three types of methyl-coenzyme M reductase from Methanococcales and Methanobacteriales.
Wagner T1, Wegner CE2, Kahnt J1, Ermler U3, Shima S4.












