Regulation of chromosome replication & cell division
Fruiting body formation with spore formation are closely coupled to cell cycle progression. Moreover, while rod-shaped vegetative cells contain one chromosome after cell division, mature spores contain two chromosomes. To understand the coupling between cell cycle regulation and development, we are focusing on understanding regulation of cell division and chromosome replication and segregation.
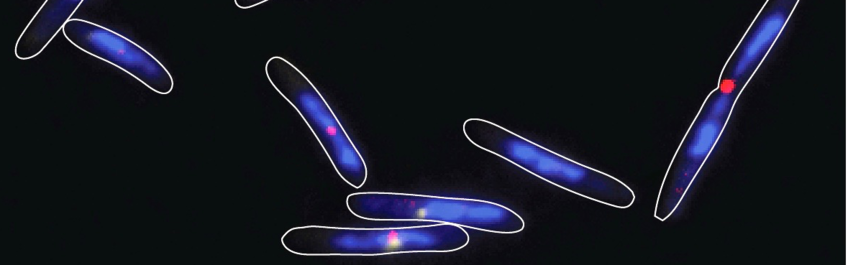
The rod-shaped M. xanthus cells divide by binary fission. We have shown that three proteins, PomX, PomY and PomZ, are important for cell division in growing cells as well as for correct positioning of the site of cell division. These effects could be traced down to the three Pom proteins being important for formation and positioning of the (Fts)Z-ring at midcell. PomX and PomY are rich in protein-protein interaction domains while PomZ is a member of the family of MinD/ParA ATPases. The PomXYZ proteins self-assemble to form a large complex that associates with the nucleoid in a manner that depends on PomZ. In addition, we found that the PomXYZ complex translocates across the nucleoid to the midnucleoid, which coincides with midcell, in a biased random walk. Once the PomXYZ complex is a midnucleoid, the complex undergoes constrained motion and stimulates Z-ring formation. During cytokinesis the PomXYZ complex splits in two and each daughter cell receives a complex that associates with the nucleoid in the daughter cells and then translocates in a biased random walk to the midnucleoid at midcell in each of the two daughters. Importantly, the PomXYZ complex localizes to midcell before as well as in the absence of FtsZ strongly suggesting that it is the PomXYZ complex that identifies the future site of cell division and then recruits FtsZ to this site.

To understand how the local protein/protein/DNA interactions in the PomXYZ system are converted into a global cellular positioning system, we searched for experimentally-based mechanisms that would give rise to a biased random walk of the PomXYZ complex to midnucleoid and constrained motion at midnucleoid at midcell. To this end, we combined experimental work and theory and developed a mathematical model that recapitulates the in vivo behavior of the PomXYZ system and explains how the PomXYZ cluster localizes to midcell. In this one-dimensional model, the diffusive flux of nucleoid-bound PomZ into the PomXYZ complex from either side of the cluster scales with the length of the nucleoid to the left and right side of the complex. A difference in the two PomZ fluxes into the cluster region results in a local PomZ concentration gradient across the cluster. The magnitude of this concentration gradient scales with the asymmetry of the cluster position on the nucleoid. Assuming that the complex preferentially moves up the PomZ gradient then the PomXYZ complex will have a bias for translocation in the direction facing most of the nucleoid. If the cluster is at the midnucleoid, which coincides with midcell until the chromosomes have segregated, the PomZ flux is equal from either side of the cluster, therefore there will be no PomZ concentration gradient over the cluster, no bias in the movements, and the motion of the cluster is constrained to midcell. Thus, in this model, the difference in the diffusive PomZ flux into the cluster from either side is a proxy for PomXYZ cluster asymmetry on the nucleoid and converts the global intracellular asymmetry of the PomXYZ cluster into a local PomZ concentration gradient over the cluster and, therefore, biased or unbiased motion. These analyses support a novel mechanism for how bacteria identify the site of cell division at midcell and nicely explains how interactions at the molecular scale are transformed into cellular organization at the µm scale.
Lately, we have also determined the spatial organization of the 9.1 Mb M. xanthus chromosome as well as chromosome dynamics during replication and segregation in vegetative cells. For chromosome segregation, M. xanthus uses a ParABS system. The genome is organized about a longitudinal axis with ori in a subpolar region and ter in the opposite subpolar region. Upon replication, one ori remains at the original subpolar region while the second copy in a directed and ParABS-dependent manner segregates to the opposite subpolar region followed by the rest of the chromosome. In parallel, ter relocates from a subpolar region to midcell. Replication involves replisomes that track independently of each other from the ori-containing subpolar region towards ter. In M. xanthus the dynamics of chromosome replication and segregation combine features from previously described systems leading to a novel spatiotemporal organization pattern.

In our current research, we focus on understanding how the PomXYZ proteins interact and how the stimulate Z-ring formationidentifies midcell. Also, we are aiming to understand how cell division and chromosome dynamics are regulated during the sporulation process.
Some of our recent publications on regulation of chromosome replication & cell division:
Lin, L., Valeriano, M.O., Harms, A., Søgaard-Andersen, L. & Thanbichler, M. (2017)
Bactofilin-mediated organization of the ParABS chromosome segregation system in Myxococcus xanthus. Nat. Comm. 8, 1817. doi: 10.1038/s41467-017-02015-z
Schumacher, D. & Søgaard-Andersen, L (2017)
Regulation of cell polarity in motility and cell division in Myxococcus xanthus. Annu. Rev. Microbiol. 71, 61-78. doi: 10.1146/annurev-micro-102215-095415
Schumacher, D., Bergeler, S., Harms, A., Vonck, J., Huneke-Voigt, S., Frey, E. & Søgaard-Andersen, L. (2017)
The PomXYZ proteins self-organize on the bacterial nucleoid to stimulate cell division. Dev. Cell 41, 299-314. doi: 10.1016/j.devcel.2017.04.011
Treuner-Lange, A. & Søgaard-Andersen, L. (2014)
Regulation of cell polarity in bacteria. J. Cell Biol. 206, 7-17. doi: 10.1083/jcb.201403136
Harms, A., Treuner-Lange, A., Schumacher, D. & Søgaard-Andersen, L. (2013)
Tracking of chromosome and replisome dynamics in Myxococcus xanthus reveals a novel chromosome arrangement. PLoS Genet. 9, e1003802. doi: 10.1371/journal.pgen.1003802
Treuner-Lange, A., Aguiluz, K., van der Does, C., Gómez-Santos, N., Lenz, P., Harms, A., Schumacher, D., Hoppert, M., Kahnt, J., Muñoz-Dorado, J. & Søgaard-Andersen, L. (2013).
PomZ, a ParA-like protein, regulates Z-ring formation and cell division in Myxococcus xanthus. Mol. Microbiol. 87, 235-253. doi: 10.1111/mmi.12094

