Bacterial adaptation and differentiation
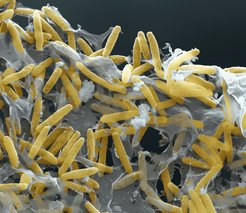
M. xanthus cells self-organize into two distinct biofilms. In the presence of nutrients, the motile, rod-shaped cells grow and divide, and if present on a solid surface, they form cooperatively feeding colonies in which cells at the edge spread outwards. In response to starvation, growth ceases, cells change their motility behavior and a developmental program is initiated that culminates in the formation of multicellular spore-filled fruiting bodies. At 24 hrs of starvation, nascent fruiting bodies have formed and the cells that have accumulated inside fruiting bodies differentiate to diploid spores. Thus, fruiting body formation is a perfect model system to analyze how bacteria coordinate changes in motility behavior, gene expression and cell cycle regulation in response to an environmental stress signal. M. xanthus belongs to the myxobacteria that are the only bacteria that cope with starvation with the formation of fruiting bodies. Amazingly, the fruiting bodies formed by different myxobacteria have very different shapes varying from haystack-shaped (as in the case of M. xanthus), stalked, coral-shaped to tree-shaped.
The overall goal of our research is to understand how M. xanthus responds to starvation with the formation of spore-filled fruiting bodies. To this end, we study at the molecular and cellular level
- intercellular communication
- signaling by the nucleotide-based second messenger c-di-GMP
- motility and its regulation
- cell cycle regulation
- Fruiting body morphology & evolution
We use a suite of experimental techniques including molecular genetics, biochemistry with protein purification and their in vitro characterization, structural biology, cell biology with live cell-imaging, proteomics, de novo sequencing of myxobacterial genomes, comparative genomics, and transcriptomics using RNAseq. Moreover, we collaborate closely with theory to develop models for the different processes that we study.
.
Intercellular communication
Over the past two decades the perception of bacterial cells as autonomous individuals each following their own agenda and not interacting with each other has been replaced by the view that bacteria interact extensively both within and between species by means of intercellular signal molecules. Each of these signal molecules constitutes part of an information processing system that is constructed of four parts: The donor cell synthesizing the signal, the signal molecule, the recipient cell, and the output response. Like in any other information processing system, the signal must be tailored to the talents of the recipient. A clear example of a tailor-made signal molecule is the intercellular C-signal in M. xanthus. Most intercellular signals in bacteria are small, freely diffusible, and part of quorum sensing systems, which helps bacterial cells to assess population size. However, that is not the case for the intercellular C-signal in M. xanthus. This signal is a 17 kDa cell surface-associated protein and non-diffusible, and C-signal transmission depends on direct contacts between cells. p17 is generated by proteolytic cleavage of the p25 precursor by the secreted protease PopC. We recently found that PopC is translocated across the outer membrane by a novel secretion system that incorporates a TonB-dependent transporter and ExbB/D homologs in the inner membrane. To understand the mechanism of the C-signal our current research focuses on understanding how PopC and p25 secretion is regulated and the identification of the p17 receptor.

Signaling by the nucleotide-based second messenger c-di-GMP
c-di-GMP is a highly versatile second messenger that is found widespread in bacteria and controls a multitude of different processes. We have shown that c-di-GMP regulates motility in M. xanthus, is essential for fruiting body formation and sporulation, and may also regulate nucleoid compaction. In our current research we are interested in identifying the enzymes that make and break c-di-GMP as well as the effector proteins that bind c-di-GMP.

Motility
Motility is essential for adaptation, adhesion to surfaces and biofilm formation in many bacteria. M. xanthus has two motility machineries that are both required for formation of spreading colonies and spore-filled fruiting bodies. One system depends on type IV pili (T4P) and cell surface polysaccharides. The second motility machinery is for gliding motility and involves the Agl/Glt motility complexes. The rod-shaped M. xanthus cells move in the direction of their long axis with clearly defined leading and a lagging cell poles. T4P are surface structures that are found on a large number of different bacterial species and they have important function in biofilm formation, virulence and motility. T4P function depends on the assembly of a macromolecular complex consisting of 14 proteins that localize to the outer membrane, periplasm, inner membrane and cytoplasm. Using cryo-electron tomography, we have recently unraveled the overall structure of architecture of this machinery. In our current research, we are testing our structure of the T4P machinery. Moreover, we are interested in understanding the structure of the gliding motility complexes. We are also interested in the biosynthetic pathways involved in synthesis of cell surface polysaccharides.
Regulation of motility & cell polarity
Motility in M. xanthus depends on two polarized motility machineries that assemble at the leading cell pole. Occasionally, cells reverse their direction of movement in a process that is stimulated by the Frz chemosensory system. During a reversal, the old leading pole becomes the new lagging cell pole and, therefore, after a reversal, the two motility machineries assemble at the new leading cell pole. Thus, the motility machineries can assemble at both cell poles but at any point in time, they only assemble at one of the poles. We are interested in understanding how this dynamic polarity of the two motility systems is regulated. In or previous work we have shown that a so-called polarity module that is composed of the four proteins MglA, MglB, RomR and RomX regulates cell polarity. All four proteins are polarly localized and with the small Ras/Rho-like GTPase being the key protein that stimulates assembly of both motility machineries. The three remaining proteins regulate the activity and localization of MglA. In our current research we are aiming at understanding how MglA, MglB, RomR and RomX localize to the cell poles and to understand in details how the Frz system interfaces with MglA/MglB/RomR/RomX.
Cell cycle regulation
Fruiting body formation with spore formation are closely coupled to cell cycle progression. While the rod-shaped vegetative cells contain one chromosome after cell division, mature spores contain two chromosomes. To understand the coupling between cell cycle regulation and development, we focus on understanding regulation of cell division and chromosome replication and segregation. The rod-shaped M. xanthus cells divide by binary fission. We have recently identified a novel cell division regulatory system, which is composed of the three proteins PomX, PomY and PomZ. The PomX/Y/Z proteins self-assemble to form a large complex that associates with the nucleoid in a manner that depends on PomZ. In addition, we found that the PomX/Y/Z complex translocates across the nucleoid to the midnucleoid, which coincides with midcell, in a biased random walk. Once the PomXYZ complex is a midnucleoid, the complex undergoes constrained motion and stimulates FtsZ-ring formation and cell division. We are also interested in understanding the spatial organization of the large 9.1 Mb M. xanthus chromosome as well as chromosome dynamics during replication and segregation in vegetative cells. In our current research, we focus on understanding how the PomX/Y/Z proteins interact and how they stimulate FtsZ-ring formation and identifies midcell. Also, we are aiming to understand how cell division and chromosome dynamics are regulated during the sporulation process.

Fruiting body morphology & evolution
M. xanthus has emerged as the model organism to understand the molecular mechanisms underlying fruiting body formation in Myxobacteria. Most Myxobacteria tested initiate fruiting body formation in response to starvation suggesting that the last common ancestor of the Myxobacteria harbored a genetic program for fruiting body formation and that fruiting Myxobacteria would share in common a genetic program underlying fruiting body formation. We used comparative and functional genomics on four complete genomes of fruiting Myxobacteria (M. xanthus, S. aurantiaca, Sorangium cellulosum and Haliangium ochraceum) and one genome of the only known non-fruiting Myxobacterium (Anaeromyxobacter dehalogenans) to test this hypothesis. Surprisingly, these comparative analyses strongly indicate that the genetic programs for fruiting body formation in M. xanthus and S. aurantiaca are very similar but significantly different from the genetic program directing fruiting body formation in S. cellulosum and H. ochraceum. In other words, our analyses reveal an unexpected level of plasticity in the genetic programs for fruiting body formation in the Myxobacteria and suggest that the genetic program underlying fruiting body formation in different Myxobacteria is not conserved. The differences in these genetic programs may either reflect convergent evolution, i.e. fruiting bodies are not homologous structures, or divergent evolution, i.e. fruiting bodies are homologous structures generated from non-homologous proteins. To follow up on these surprising findings, we are currently generating high quality, completed genome sequences of selected Myxobacteria. Moreover, we conduct comparative transcriptomics analyses to map similarities and differences between transcriptional programs in different Myxobacteria during fruiting body formation.














