Microbial Networks

A major goal of our research is to quantitatively understand the molecular and physiological functions of cellular networks in microorganisms. As model systems, we study several networks that regulate key cellular processes and behaviors in bacteria and yeast, primarily using the well-established model organisms E. coli and S. cerevisiae. We apply a variety of fluorescence microscopy techniques, molecular biology, transcriptomics, proteomics, metabolomics, and biochemistry to investigate the ability of protein networks to sense and integrate multiple stimuli and reliably control cell behavior and physiology. We also aim to better understand the relationship between single-cell and collective behaviors within microbial communities, including cell differentiation and the role of chemical communication and physical interactions. Finally, we use experimental evolution to gain insights into the evolvability of cellular functions and microbial communities. In most of our work, we combine experiments with theoretical analysis and computational modeling to elucidate principles common to different biological systems and to understand the physics behind biology. Finally, we explore how cellular functions can be redesigned for potential bioengineering applications.
Environmental sensing and signal processing by bacterial chemotaxis networks
Most motile bacteria are capable of following gradients of various chemical and physical stimuli, such as nutrients, signaling molecules, pH, temperature, osmolarity, and harmful chemical agents in their environment. This tactic behavior is highly advantageous for bacterial survival and competition for nutrients, but also plays an important role in the formation of multicellular communities and the colonization of animal and plant hosts. We study how the chemotaxis network of E. coli and other bacteria senses, processes and integrates multiple environmental cues encountered in animal hosts and aquatic or terrestrial habitats. We use biochemical assays for high-throughput identification of chemoeffectors for different bacterial species, including animal pathogens and commensals. We complement this with Förster resonance energy transfer (FRET) measurements of pathway activity and microscopic analyses of bacterial behavior in microfluidic channels to characterize the bacterial response to these stimuli. Finally, we investigate the physiological and ecological relevance of bacterial motility and chemotaxis in different habitats, including the animal gastrointestinal tract and the rhizosphere.
Bacterial motility and chemotaxis as a model for evolutionary optimization.
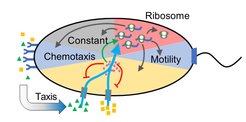
Microbes are excellent models for addressing fundamental biological questions. One such question is whether evolution has optimized the function of cellular networks and whether we can understand the logic of this optimization. We are currently investigating the regulatory rules that govern the allocation of protein resources to bacterial motility. Since swimming is one of the costliest cellular functions, investment in motility gene expression must be tightly controlled. We have shown that the expression of motility genes in E. coli is anticipatory, proportional to the potential benefit of finding additional sources of nutrients, but also limited by physics of bacterial swimming. Moreover, we observed that the motility of many natural clinical isolates of E. coli is optimized for swimming in a porous medium, apparently reflecting the environment in the animal gut.
We also use experimental evolution to investigate how bacterial motility and chemotaxis are tuned under selection. We observed that the E. coli chemotaxis pathway can recover its functionality upon loss of normally essential chemotaxis genes by evolving a distinctly different chemotaxis strategy, demonstrating its surprisingly high evolvability. We are currently investigating the ability of the E. coli chemotaxis pathway to evolutionarily accommodate genes from other bacterial species, as occurs naturally during lateral gene transfer, and what defines the limits of this adaptability.
Ni, B., Colin, R., Link, H., Endres, R.G., and Sourjik, V. (2020). Growth-rate dependent resource investment in bacterial motile behavior quantitatively follows potential benefit of chemotaxis. Proc Natl Acad Sci USA 117, 595-601 Laganenka, L., Colin, R., and Sourjik, V. (2020). Flagella-mediated mechanosensing and RflP control motility state of pathogenic Escherichia coli. mBio, 11(2), e02269-19
Regulation of transition between motile and sessile lifestyle
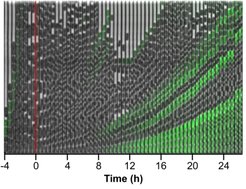
Activation of the general stress response and biofilm formation occurs in bacteria prior to entry into the stationary phase of growth and is tightly linked to the regulation of motility. We are therefore interested in understanding how this major transition in bacterial lifestyle is controlled at the regulatory level. We have recently shown that this transition is not uniform in the E. coli population, but rather results in the emergence of two distinct subpopulations of cells that are either committed to biofilm formation or not. We are dissecting the highly complex regulatory network that controls this transition, including the regulatory interplay between motility and biofilm genes and second messenger signaling. As a part of this research, we have established a set of FRET-based reporters to follow the real-time dynamics of the second messenger c-di-GMP, which governs this transition, in single growing cells. Finally, we have recently uncovered several new mechanisms of motility gene control in E. coli, including those specific to pathogenic E. coli isolates, and are investigating the molecular details of these mechanisms.
Biochemical noise and dynamics in cellular networks.
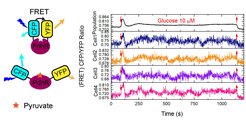
Biochemical reactions in cellular networks are inherently stochastic, which can lead to random vfluctuations in network activity. Such noise can have profound effects on the ability of networks to reliably perform their functions, but can also result in beneficial variability among cells in a population. While stochasticity in gene expression in microorganisms has received much attention, the extent and importance of post-translational biochemical noise in cellular networks remains largely unknown. We use the FRET assay to monitor signaling and metabolic networks in single E. coli cells, revealing surprisingly large and dynamic fluctuations in network activity under some conditions. We aim to combine these measurements with mathematical modeling to elucidate the origins and physiological consequences of biochemical fluctuations.
We have also systematically quantified mobility, a fundamental biophysical parameter that constrains network dynamics, for a large set of proteins in the E. coli cytoplasm by combining fluorescence correlation spectroscopy (FCS) with computer simulations and mathematical modeling. We also study the dependence of protein diffusion on cell growth and various perturbations, including antimicrobial treatments.
Dynamics and evolution of mutualistic microbial consortia
Many microorganisms exist within communities that rely on the exchange of metabolites for their growth. However, although such mutualistic interactions may be common in nature, their determinants, emergence and evolution towards full-fledged symbiosis remain poorly understood. To address these questions, we established a synthetic mutualistic community between a eukaryotic (S. cerevisiae) and a prokaryotic (E. coli) microorganism. We used this consortium to demonstrate the importance of physical association between community members and motility for community stability. We are currently proceeding with the experimental evolution of this consortium under selective conditions and have demonstrated that emergence of increased metabolic entanglement between partners can be experimentally reproduced and studied at the molecular level under laboratory conditions.
Recent publications:
Scarinci, G., Sourjik, V. (2022) Impact of direct physical association and motility on fitness of a synthetic interkingdom microbial community. The ISME journal
Chemotactic bacteriabots and minicells for cargo delivery

We are exploring the use of chemotactic bacteria to deliver the microscopic cargo. We have developed a system for rapidly coupling microscopic cargo to motile E. coli cells and demonstrated that the resulting biohybrid bacteriabots remain motile and chemotactic, i.e., capable of transporting microscopic cargo in chemical gradients. In addition to using regular E. coli cells, we have also developed the production of engineered motile and chemotactic E. coli minicells. These minicells are chromosome-free but can carry and express proteins, making them attractive tools for pharmaceutical applications such as drug delivery
Transport of microscopic cargo by a bacteriabot
Bacteriabot based on elongated (cephalexin-treated) motile E. coli cell, transporting a 2.2-µm polymethyl methacrylate (PMMA) microparticle.
Recent publications:
Ni, B., Colin, R., Sourjik, V. (2021) Production and Characterization of Motile and Chemotactic Bacterial Minicells, ACS Synth Biol.;10(6):1284-1291.
Pheromone communication in yeast mating.
We study the principles of pheromone communication during mating of the unicellular eukaryotic microbe S. cerevisiae as a simple model of mating behavior and cellular information processing. We investigated how this information is reliably transmitted by the MAPK signaling pathway in the presence of cellular noise. We also aim to better understand the information is provided by the mating pheromones and the physiological reasons for the asymmetry between the pheromones secreted by two different mating types of S. cerevisiae. We were able to show that this asymmetry in signaling is reflected in mating behavior, which has important implications for understanding early steps in the evolution of sexual dimorphism in eukaryotic organisms, one of the major evolutionary transitions in biology.
Recent publications:
Anders, A., Colin, R., Banderas, A., Sourjik, V. (2021) Asymmetric mating behavior of isogamous budding yeast. Sci Adv, 7(24):eabf8404.








