
Establishment of multiplex CRISPR-Cas9 genome editing in Ustilago maydis
The adoption of CRISPR-Cas9 genome editing system in U. maydis allowed not only the efficient disruption of single genes but also opened the door for the simultaneous disruption of genes harboring identical target sequences. Taking advantage of the nature of the system, a sgRNA can be designed to target a conserved region present in several genes. In U. maydis, there are only a handful of effectors that share stretches of identical sequences that can be disrupted in this way. We tested this possibility targeting a conserved region of two effector genes of the eff1 effector family. In this case, both genes are located next to each other in the genome and the induced editing resulted in a chromosomal deletion of the region between the cleavage sites in all cases.
To inactivate members of gene families which lack stretches of sequence identity several sgRNA needed to be expressed simultaneously with the Cas9 protein. We opted for an all-in-one-plasmid strategy and modified our Cas9 expression plasmid to harbor several sgRNA expression cassettes (Figure 6). The sgRNA expression cassettes consist of a tRNA promoter and a sgRNA and can be cloned in to the Cas9 expression plasmid easily using Gibson assembly. Since the efficiency of Cas9 mediated genome editing in a multiplexing construct might decrease due to the competition of different sgRNAs for the Cas9 proteins, we increased the expression of Cas9 by inserting a stronger promoter and implemented an additional selection and incubation step after transformation to increase the time of action of Cas9 in the cells. As a proof of concept we generated a multiplexing construct targeting five genes belonging to the eff1 effector family. Following the modified protocol, 70% of the tested transformants displayed gene editing events in all five genes. The same approach is now used to inactivate all core effector genes which reside in gene families.
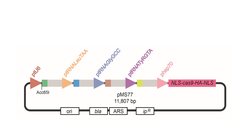
Figure 6: Schematic representation of the plasmid constructed for targeting several genes by Cas9 in U. maydis.
