Discovery of circular RNA molecules involved in protein trafficking
Signal recognition particles (SRPs) are found in all living organisms on earth. They contain a short RNA molecule which is essential for cellular trafficking and secretion of proteins. Scientists in the research group headed by Dr. Lennart Randau at the Max Planck Institute for Terrestrial Microbiology in Marburg in their recent publication in the journal eLife describe the discovery of a novel type of SRP RNA molecules that are circularized to function in the cell. These unusual RNA circles are generated by a moonlighting activity of the machinery that is usually responsible for the removal of introns in transfer RNAs.
All cells on earth generate proteins that need to be transported to the cell membrane (in prokaryotes) or the endoplasmic reticulum (in eukaryotes) to allow their secretion. These proteins contain a signal peptide sequence that is recognized by the essential and evolutionarily conserved SRP pathway. A SRP is always a ribonucleoprotein complex consisting of a single RNA molecule and few associated proteins. However, genome sequencing of archaeal species of the genus Thermoproteus did not identify the standard SRP RNA gene. ”The observation that these archaea do not contain a standard SRP RNA was so surprising to us that we wanted to solve the mystery of the “missing” component of this essential protein-targeting machine in these organisms” explains Andre Plagens, the first author on the paper.
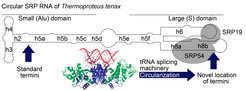
Using their knowledge about small RNA processing pathways and a combination of next-generation sequencing, biochemical and microbiological methods lead to the discovery that the “missing” universal SRP RNA gene was not yet identified due to its permutation; i. e. rearrangements of the genome resulted in a swap of the left and right portions of the SRP RNA gene. Remarkably, the correct sequence order is restored in the mature SRP RNA molecules when their ends become permanently linked in a circular SRP RNA.
This unexpected discovery of circular SRP RNAs immediately raised additional questions: How are the RNA circles formed? And, why is the SRP RNA gene fragmented? The first question was answered quickly. It was shown that the machinery that removes introns from transfer RNAs is responsible for SRP RNA circularization. This result highlights that moonlighting activities can evolve from ancient pathways.
The second question shines a light on the evolution of genome fragmentation. Circular RNA molecules are very stable and that might be an advantage for Thermoproteus species which grow at temperatures of up to 95 °C. It is possible that the described reaction can be utilized to stabilize different synthetic RNA molecules. Future studies will also investigate the selective pressure that favors the evolution of fragmented non-coding RNA genes. Viruses that specifically use these genes as attachment sites might play a crucial role.
These findings were described in a recent study that appeared in the journal eLIFE:
Plagens, A., et al. eLIFE (2015) doi: http://dx.doi.org/10.7554/eLife.11623).
http://elifesciences.org/content/early/2015/10/24/eLife.11623
Contact: Dr. Lennart Randau (lennart.randau@mpi-marburg.mpg.de)
To learn more about Dr. Randau’s research please visit: http://www.mpi-marburg.mpg.de/randau/





![<p>Discovery of [Fe]-hydrogenase in bacteria opens new possibilities for conversion of hydrogen</p>](/506336/original-1555497672.jpg?t=eyJ3aWR0aCI6MzYwLCJoZWlnaHQiOjI0MCwiZml0IjoiY3JvcCIsImZpbGVfZXh0ZW5zaW9uIjoianBnIiwib2JqX2lkIjo1MDYzMzZ9--d3d7c57e064104c2acc573916156127d6bedd85b)


![Protection of [Fe]-hydrogenase](/416905/teaser-1561027823.jpg?t=eyJ3aWR0aCI6MzYwLCJoZWlnaHQiOjI0MCwiZml0IjoiY3JvcCIsImZpbGVfZXh0ZW5zaW9uIjoianBnIiwib2JqX2lkIjo0MTY5MDV9--6ea496a03c86ba3d98a19e05aac63809917fe800)



