Small proteins in bacterial signaling network
Regulation of the PhoQ sensor kinase by small proteins
Central to the PhoQ/PhoP two-component signaling pathway, the PhoQ sensor kinase determines the sensitivity and output of the system and acts as a master regulator of the bacterial virulence program. PhoQ is associated with small proteins that modulate bacterial virulence, fitness, and drug resistance in E. coli and other pathogenic enterobacteria (Fig. 1).
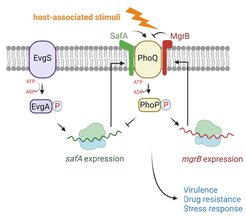
Fig. 1. PhoQ and its regulatory small proteins. The sensor kinase PhoQ detects host-associated stimuli, such as antimicrobial peptides, low Mg2+, low pH, and osmolarity upshift, which leads to an upregulation of genes involved in bacterial virulence, drug resistance, and stress responses. Two small proteins directly interact and regulate PhoQ kinase activity in E. coli. The small protein MgrB inhibits PhoQ, and SafA activates it. The expression of mgrB is regulated by the PhoQ/PhoP two-component system. The expression of safA is regulated by the EvgS/EvgA two-component system.
MgrB and SafA are small membrane proteins with a single transmembrane helix. MgrB inhibits PhoQ and SafA activates it in E. coli (Fig. 1). However, how bacteria coordinate these opposing regulations is unknown. Do MgrB and SafA compete for PhoQ binding or form a tripartite complex? Is MgrB as stable as SafA? Does SafA bind to PhoQ with the same affinity as MgrB, and does this affinity change over the course of infection? How does the coordinated regulation affect bacterial virulence? Combining genetic, molecular, and cellular approaches, we investigate the regulation by MgrB and SafA in the context of mammalian cell infection by pathogenic E. coli.
Engineering new antimicrobials targeting sensor kinases
Many bacterial sensor kinases sense environmental changes, control virulence, and are essential for bacterial survival and host infection. Sensor kinases are ideal targets for developing specific and selective antimicrobials, as they have structurally unique sensor domains, are prevalent in prokaryotes, and are absent in the animal kingdom. As sensor kinases are not essential for bacterial survival, targeting specific sensor kinases does not result in high selection pressure, thus avoiding rapid mutation and adaption, leading to drug resistance.

Fig. 2. Schematic representation of MgrB mediating PhoQ to sense AMPs. PhoQ dimer (green and gray) is active under physiological magnesium conditions, inducing mgrB expression. MgrB can then bind to PhoQ, resulting in a partially repressed state. AMPs enter the bacterial periplasm, bind to PhoQ, and cause MgrB to dissociate from PhoQ. The figure is adapted from Jiang et al., PNAS (2023).
We elucidated the inhibitory mechanism of MgrB on the sensor kinase PhoQ and found that the binding of MgrB to PhoQ is reversible. Low amounts of antimicrobial peptides initiate the dissociation of MgrB from PhoQ, which leads to activation of the PhoQ/PhoP system and increased drug resistance. Therefore, MgrB acts as a bacterial sensor to detect subinhibitory concentrations of antimicrobial peptides (AMPs) (Fig.2). Utilizing the knowledge we gained from MgrB, we aim to develop SuperMgrBs (strong inhibitors that bind to PhoQ with high affinity and inhibit its function irreversibly). SuperMgrBs are expected to fix PhoQ in an inactive conformation, repress bacterial virulence program, and reduce drug resistance. Additionally, with a collaborative effort, we attempt to generate de novo PhoQ inhibitors via structural, computational, and genetic approaches.
Function discovery of small membrane proteins
Numerous small proteins have been discovered in three domains of life, among which a significant portion is predicted to localize to the membrane. Based on a few well-studied examples, small membrane proteins are regulators involved in various biological processes ranging from bacterial virulence regulation to immunosurveillance in human. However, the function of most newly discovered small membrane proteins awaits validation or remains elusive. Their small size and hydrophobicity make protein production challenging, hindering function discovery.
We combined a defined cell-free system with lipid sponge droplets and synthesized small membrane proteins in vitro. Lipid sponge droplets contain a dense network of lipid bilayers, which accommodates and extracts newly synthesized small membrane proteins from the aqueous surroundings (Fig. 3 and Movie 1). With this system, we produced small membrane proteins up to micromolar scale sufficient for downstream functional analyses. The produced small proteins are also suitable for identifying their interacting partners. We are interested in using this powerful tool to explore the function of newly discovered small membrane proteins in E. coli and other enteric pathogens.

Fig. 3. The synthesis of mNeonGreen-MgrB using a cell-free system with lipid sponge droplets. Representative images collected in the transmitted light (TL), GFP, and merged channels are shown in the left, middle, and right panels. The figure is adapted from Jiang et al., Biorxiv (2023).


