Motility allows Adaptation
Study shows how pathogenic bacteria can adapt to varying conditions of the digestive tract
Basic, acidic, basic again: for pathogenic bacteria such as Salmonella, the human digestive tract is a sea change. So how do the bacteria manage to react to these changes? A team of researchers led by Andreas Diepold has now provided a possible explanation: pathogenic bacteria can change components of their injection apparatus on the fly - like changing the tires on a moving car - to enable a rapid response.

In order to increase survival chances in contact to eukaryotic host cells, both symbiotic and pathogenic bacteria have developed methods to influence host cell behavior. The type III secretion system (T3SS) injectisome is a molecular machinery used by various pathogenic bacterial genera, including Salmonella, Shigella, pathogenic Escherichia, Pseudomonas, and Yersinia to deliver molecular toxins—effector proteins—directly into the eukaryotic host cells. Researchers in the laboratory of Andreas Diepold at the MPI for Terrestrial Microbiology have now found that high dynamics of this apparatus allow bacteria to adapt quickly to changing conditions in the digestive tract.
Some of the best-known human pathogens - from the plague bacterium Yersinia pestis to the diarrhea pathogen Salmonella - use a tiny hypodermic needle to inject disease-causing proteins into their host's cells, thereby manipulating them. This needle is part of the so-called type III secretion system (T3SS), without which most of these pathogens cannot replicate in the body.
Only recently it was discovered that large parts of the T3SS are not firmly anchored to the main part of the system, but are constantly exchanging during function. However, the significance of this phenomenon remained unclear. Researchers in the laboratory of Andreas Diepold at the MPI for Terrestrial Microbiology have now discovered that this dynamic behavior allows the bacteria to quickly adapt the structure and function of the injection apparatus to external conditions.
The digestive system: an alternating bath for bacteria
Human digestion starts with a neutral to slightly alkaline environment in the mouth and esophagus, which the addition of gastric acids suddenly changes to strongly acidic in the stomach - an environment that many pathogens do not survive. The actual target of Yersinia enterocolitica, the pathogenic bacteria investigated in the study, is the intestine. Here, pH-neutral conditions are restored.
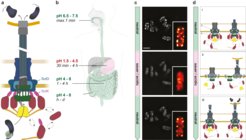
Adaptation of the bacterial type III secretion system to changing pH. (a) Schematic representation of the active T3SS injectisome. Effector proteins (black/blue) are exported from the bacteria to the host cell in a single transport step. The position of the pH sensor protein SctD and one of the dynamic cytosolic T3SS components, SctK, are indicated. Double arrows indicate the exchange of the cytosolic subunits or subcomplexes between the cytosolic and the injectisome-bound state. (b) pH ranges and typical retention times at different parts of the gastrointestinal system. (c) Fluorescent micrographs of live Y. enterocolitica expressing a fluorescently labeled cytosolic T3SS component (EGFP-SctK). Images were taken 10 minutes after bacteria were subjected to the indicated pH. Insets, enlarged single bacteria in “red-hot” color scale. Scale bar, 2 µm. (d) Model of the pH-dependent suppression of T3SS activity. From top: (i) Assembly of the T3SS upon entry into host organisms; cytosolic components bound and exchanging with cytosolic pool. (ii) Prevention of effector translocation upon host cell attachment in low pH environment, because cytosolic components are exclusively cytosolic. (iii) Re-association of cytosolic components to the injectisome and effector translocation upon host cell contact in neutral body parts. The pH sensor protein SctD and the cytosolic components SctK are indicated by D and K, respectively.
But how do the bacteria manage to adapt so quickly to the changing conditions, and how is this controlled? PhD student Stephan Wimmi, the first author of the study, was able to demonstrate that a protein in the bacteria's membrane acts as a sensor for the pH value. In a collaboration with Ulrike Endesfelder's lab at the MPI, he found that this protein becomes more motile at low (= acidic) pH and thus transmits the signal to the T3SS components inside the bacterium.
Flexibility prevents misfiring
In an acidic environment like the stomach, the mobile components do not bind to the rest of the apparatus (including the needle itself), so that the injection system remains inactive. As soon as the bacteria enter a pH-neutral environment – as it is found in the intestine -, the dynamic proteins reassemble, so that the T3SS can quickly become active at these sites - to the possible distress of the infected person.
The researchers speculate that the newly discovered effect may allow the bacteria to prevent an energy-consuming "misfiring" of the secretion system in the wrong environment, which could even activate the host's immune response. On the other hand, the mobility and dynamics of the structure allows the system to be rapidly reassembled and activated under appropriate conditions.
Protein mobility and exchange are increasingly being discovered in complexes and nanomachines across all domains of life; however, the utility of these dynamics is mostly not understood. The new results from Marburg, published in the journal Nature Communications, show how protein exchange allows to respond flexibly to external circumstances - an immense advantage, not only for bacteria.













