
Prokaryotic Small RNA Biology
Our research is aimed at understanding the processing of small RNAs involved in the defense against integrative elements (e.g. viruses) in Bacteria and Archaea. We investigate the RNAs and ribonucleoproteins that play the central role in the prokaryotic CRISPR (Clustered Regularly Interspaced Short Palindromic Repeats) immune system. Second, we study the evolution of diverse disruptive elements within archaeal non-coding RNA genes. These systems will be exploited for the modulation of prokaryotic immunity and the creation of gene knock-down technology. Our research uses an interdisciplinary approach combining computational, biochemical and microbiological techniques.
CRISPR RNA processing
The presence of CRISPR arrays in many bacterial and most archaeal genomes serves as molecular evidence for their ongoing battle with integrative elements. The Clustered Regularly Interspaced Short Palindromic Repeats (CRISPR) RNAs play a central role in this prokaryotic immune system. CRISPR elements were shown to confer resistance against viruses and to prevent conjugation and plasmid transformation. Each CRISPR element is composed of direct repeats separated by similarly sized non-repetitive spacers. Many CRISPR spacer elements are derived from viral sequences and provide insights into the changing host/virus relationship during evolution. In 2007, Barrangou and colleagues showed that the presence of a virus in the growth medium can lead to the acquisition of a new spacer within the host DNA containing a small sequence fragment of this specific virus. Once a new spacer is inserted into the host's CRISPR element, the host is resistant against the attacking virus.
Archaea
Bacteria and Eukaryotes (including animals, plants and fungi) are only two of the three domains in the tree of life. In 1990, Carl Woese, Otto Kandler and Mark Wheelis utilized molecular evidence including ribosomal RNA based phylogenies to propose the divide of the morphologically similar prokaryotes into Bacteria and the new third domain, the Archaea. Like Bacteria, Archaea are also single-celled microorganisms. However, they exhibit many features and metabolic pathways that indicate their close evolutionary relationship with the Eukaryotes. Most notably, these features include their transcription and translation machineries. Certain biochemical aspects, like the unique cell membrane composition and the great variety of usable energy sources, are unique to the Archaea.
Archaea exist in many different habitats, including oceans and soils, but can also be found in extreme environments where they survive high temperatures or highly saline, acidic or alkaline conditions. One example is the archaeon Methanopyrus kandleri which grows at up to 122 °C, a temperature reached in the fluid of black smokers at the bottom of the ocean.
Our group is interested in RNA processing pathway in Archaea. We discovered several unusual pathways for the processing of transfer RNAs and we will continue to investigate the evolution of these RNA processing machineries. Furthermore, Archaea exhibit the greatest variety of CRISPR systems and we will use our knowledge obtained in studying tRNA processing to investigate the processing of the small RNAs involved in this exiting defense system against integrative elements like viruses or conjugative plasmids.
Transfer RNA splicing in Archaea
Transfer RNAs are the well-characterized adaptor molecules which mediate the recognition of codons and their translation into the appropriate amino acid during ribosomal protein biosynthesis. To be able to fulfill this crucial cellular function, all canonical tRNAs show a conserved cloverleaf-shaped secondary structure and fold into an L-shaped tertiary structure. They are short RNA strands with an average length of 76 nucleotides. It would be logical to assume that such small RNA molecules could be encoded by short continuous genes. This is true for most tRNAs, yet, most notably in Archaea, a surprisingly diverse repertory of creatively disrupted tRNA genes exists.
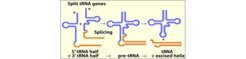
The earliest discovered disrupted tRNA genes were tRNA precursors that contain an intron sequence located one nucleotide downstream of the anticodon. Such introns are found in tRNA genes from all three domains of life, but the machineries required to remove these intervening sequences differ significantly. The archaeal machineries involve a splicing endonuclease that recognizes a characteristic structural motif (the bulge-helix-bulge BHB motif) located at the intron-exon junctions of tRNA precursors. The utilization of this motif allowed the evolution of introns at varied positions within the tRNA gene and there are also cases of tRNA genes with multiple introns. In 2005, we found that tRNA genes can be split into two tRNA half genes that contain complementary GC -rich ends which are proposed to serve as "sticky" sequences to aid the meeting and annealing of the two halves (Fig. 1). The annealed halves are then processed by a splicing endonuclease in an unusual trans-splicing process.
Why did these disrupted tRNA genes evolve and why where they maintained as a seemingly advantageous feature during evolution? We will investigate this question by analyzing the correlation of tRNA genes with viral attachment sites. Most archaeal viruses use large tRNA gene portions for their integration which would be hindered by any disruptive element within a tRNA gene.
