Bacterial Secretion Systems
Secretion systems allow bacteria to interact with other species, including eukaryotes. They are key players in bacterial virulence and competition.
My group wants to understand how secretion systems work on the molecular level, how they are activated and regulated in situ, how they contribute to bacterial virulence and competition and how different secretion systems are co-regulated and influence each other.
A focus of our research is the type III secretion system (T3SS), which bacteria use to inject effector proteins into eukaryotic target cells. This system, also called injectisome, is essential for virulnece in important human pathogens, but also plays a role in symbiosis.
We aim to uncover the molecular mechanism of protein export by the T3SS and link these findings to its physiological impact. Accordingly, we apply a wide range of methods, both in our lab and in collaborations with leading researchers within and beyond the Max Planck Institute. These range from cutting-edge live-cell and superresolution microscopy, complemented by various biochemical and genetic methods, to cell culture-based infection assays. Wherever possible, we analyze the T3SS in live bacteria, both under controlled conditions and in contact to host cells. This approach allows us to directly test for the physiological significance of our findings for T3SS function and successful infection.
Current priorities of our research are listed below.
The molecular function and dynamics of the type III secretion system
Despite years of dedicated research, the molecular events that lead to effector export through the T3SS are still unknown. The components that govern these events are located at the cytosolic interface of the injectisome. Using single-molecule tracking to monitor these components in the working T3SS in real time, we have recently discovered that parts of the T3SS are dynamic, exchanging subunits between a cytosolic pool and the injectisome, while secreting effectors (Diepold et al., 2015). Strikingly, the exchange correlates with the function of the T3SS. We found that the reason for this unexpected behavior is a switch in the interactions between the cytosolic T3SS components upon activation of the system (Diepold et al., 2017).
But how exactly does this dynamic behavior enable secretion? Does it directly allow to activate the system in response to external signals? And could this be an Achilles heel we can exploit to find a generally applicable treatment for T3SS-dependent bacterial infections?

To answer these questions, our group visualizes the T3SS and its substrates during its function in live bacteria, from the cellular to the single-molecule level. Using this approach in combination with in situ interaction studies, we recently found that cytosolic components of the T3SS provide a shuttle service for effector proteins, ensuring their specific and efficient export by the injectisome (Wimmi et al., 2024).
Working with live bacteria, we can directly compare the molecular events under different conditions. This allows us to determine the changes that occur upon external cues, such as the activation of the system in vitro or by direct host cell contact.
Activation and regulation of the T3SS during infection
Bacteria need to make sure that the right amount of the correct effector proteins is translocated into a target cell without alerting the host immune system. Therefore, substrate specificity and translocation rate of the injectisome must be tightly controlled, in acute as well as in persistent infections.
The activity of the T3SS thus needs to be regulated; however, little is known about how environmental signals regulate the function of the T3SS. Our lab has recently discovered that T3SS composition and function quickly and strongly respond to changes in the external conditions, such as the local pH. In acidic environments, the cytosolic T3SS components reversibly detach from the machinery which temporarily suppresses T3SS activity. Upon restoration of neutral pH, these components reassemble and allow the bacteria to quickly activate protein secretion at the native site of action of the T3SS (Wimmi et al., 2021), a striking example for the functional regulation of the T3SS by the environment. We now want to identify the regulatory pathways involved in this regulation, and especially the events at the injectisome itself.
To better understand how different bacteria use the T3SS, we compare its function and regulation in Yersinia enterocolitica and Pseudomonas aeruginosa. Applying our experience in live microscopy to the P. aeruginosa T3SS allows us to build on the wealth of studies that have described its regulatory networks, and to determine the link between the signaling pathways and the T3SS. Regulation of the T3SS in P. aeruginosa is directly relevant for the outcome of nosocomial infections, and hence a better understanding is a key step towards preventing healthcare-related infections. Comparing the regulation of the Y. enterocolitica and P. aeruginosa T3SS, we can discern general and species-specific mechanisms in the regulation of type III secretion, a crucial prerequisite for the development of generic T3SS inhibitors.

Controlling the T3SS in health and disease
The better understanding about the molecular function and regulation of the T3SS that we want to achieve has important practical implications, and we directly apply our research results to be able to efficiently control the function of the T3SS. By comparing regulatory principles of the T3SS of various pathogens during infection, we can identify conserved targets that allow to inhibit toxin delivery, and subsequently virulence, in many important pathogens. In addition, we use our knowledge on the regulation of the T3SS to engineer bacteria, for example for controlled protein delivery to host cells through the T3SS.
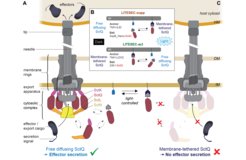
In a recent study, we exploited the dynamic nature of the T3SS to control protein secretion and translocation into eukaryotic cells by light. By combining optogenetic interaction switches with a dynamic cytosolic T3SS component, the cytosolic availability of this essential T3SS component and in consequence T3SS-dependent effector secretion can be regulated by external light. The resulting system, which we called LITESEC-T3SS (Light-induced translocation of effectors through sequestration of endogenous components of the T3SS), allows rapid, specific, and reversible activation or deactivation of the T3SS upon illumination (Lindner et al., 2020).
To make this powerful approach more widely applicable for microbiology, we adapt and improve optogenetic switches for use in prokaryotes (Lindner and Diepold, 2022).
Beyond the direct potential of such synthetic biology approaches in biotech and medicine, the ability to precisely control T3SS activity will advance our understanding of T3SS function and regulation, and therefore feed back into our core research.
Co-regulation and crosstalk of bacterial virulence factors and cell biology
To thrive in challenging and ever-changing conditions, such as inter-bacterial competition and in host organisms, bacteria encode various virulence factors. The timely deployment of these factors is often essential for survival and proliferation during infection, and spatial arrangement and temporal activity of the systems influence each other. Studying this co-regulation is essential to understand the biology of prokaryotes in isolation and especially in community with other bacteria and during infections and other interactions with eukaryotic cells. At the moment, studies of this type focus mainly on the regulation of transcription and protein expression, which can be studied globally using "omics" approaches. While such studies have provided important insights into the complex expression control of various virulence factors and other protein complexes, they cannot detect dynamic responses of the system, such as changes in protein function, interaction, and localization.
Therefore, a central goal of our work is to understand the functional co-regulation and interaction of key bacterial virulence and adaptation mechanisms under changing conditions. An optimal model system for these studies is Pseudomonas aeruginosa, an opportunistic pathogen that employs a large arsenal of virulence factors in a targeted and tightly regulated manner. Our group is studying the expression, assembly, and function of these virulence factors at the single-cell level to directly observe the direct interactions of the systems.
In addition to interacting with each other, the function of virulence factors is often tightly interwoven with bacterial cell biology. A striking example is the inhibition of bacterial growth upon activation of the type III secretion system, which led to the discovery of the system some 40 years ago, but whose basis is still poorly understood.
We recently discovered how a toxin-antitoxin system ensures the maintenance of the T3SS despite these costs (Schott et al., 2023). and currently investigate how, conversely, subtle changes in the bacterial cell biology can lead to a previously unknown specific downregulation of the T3SS.
A better understanding of these complex relationships and reciprocal influences offers a unique insight into the cellular biology of prokaryotes and the basis of bacterial survival in contact with eukaryotic host cells. Moreover, it is an essential step towards identifying targets for infection-inhibiting agents, a key goal against the backdrop of an impending post-antibiotic era.
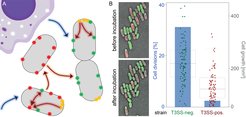
(A) Schematic representation of bacterial subpopulations harboring virulence systems (red, green and yellow foci) and their regulation by external cues (blue arrows) and crosstalk within and between bacteria (red and orange arrows, respectively). (B) Growth and division of T3SS-positive (red) and T3SS-negative (green) Y. enterocolitica on LB-agarose pads under secreting conditions over two hours. Left, fluorescence micrographs showing growth and division of green (T3SS-negative), but not of red (T3SS-positive) bacteria within the analysis period. Right, quantification of fraction of cell divisions (blue bars and axis on left side) and cell growth (increase in two-dimensional cell area on micrographs) per initial bacterium (box charts and single data points, right axis) for the indicated strains. Adapted from Milne-Davies et al., 2019.







