Complex Adaptive Traits (CATs)
in beneficial Bacilli
Bacteria have evolved sophisticated strategies to cope with stress and adapt to environmental change. Our group studies complex adaptive traits (CATs) in beneficial Bacilli at the behavioral and mechanistic level. Our research reveals new traits and aims to understand how the gene regulatory network controls them. We specifically focus on spores and peptide-based signaling networks and seek to identify fundamental organizational principles relating the molecular network design to population-level behavior and vice versa. This should facilitate rational manipulations of bacterial populations and the development of probiotics in the future. In cross-disciplinary studies together with others we also investigate stress-associated phenomena with relevance for humans and the environment (e.g. formation of methane) using B. subtilis as a model organism.
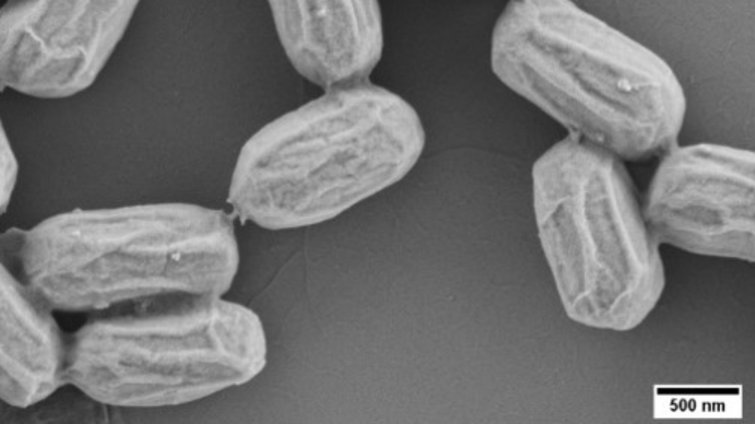
Bacillus subtilis: a spore-forming model organism with many applications
Research in our lab focuses on CATs in Bacilli for two reasons. First, these bacteria have the remarkable ability to differentiate into dormant and very resistant endospores. Spores can survive adverse conditions for many years, yet they can revive almost instantly when conditions improve. The spore is the prevalent form in which these bacteria are found in nature, and spores revive both in the soil and in the gut environment. Second, some Bacilli have positive effects on other organisms. In fact, biotechnology companies produce tons of spores annually in order to administer beneficial bacteria to plants, animals and humans, thereby reducing the use of agrochemicals and antibiotics. Most of our research has focused on the model bacterium B. subtilis. We have recently started to work also on other beneficial Bacilli, including Bacillus velezensis and Paenibacillus polymyxa.
Complex adaptive behaviors
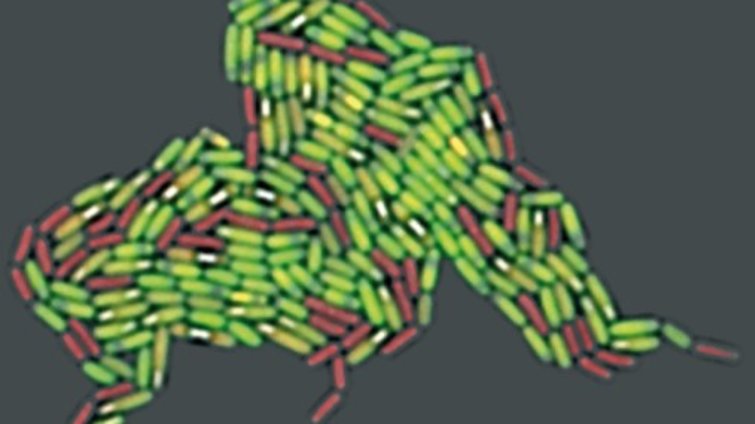
Complex adaptive traits (CATs) are still an understudied area in the field of bacterial stress responses in general, although it is well known and readily observed that bacteria diversify phenotypically under stress. In the model organism Bacillus subtilis, sporulating and replicating cells coexist for some time, the resulting spores have different properties and vegetative cells can differentiate into morphologically distinct cell types, of which only one activates the production of a beneficial secondary metabolite. All three phenomena are being investigated in our lab. Phenotypic population heterogeneity has obvious advantages for a clonal organism: Firstly, diversification is a powerful means of mitigating risks and exploiting opportunities arising from unpredictable changes in environmental conditions. Furthermore, functionally diversified populations enable the division of labor. On the other hand, heterogeneous behavior is often undesirable in biotechnical applications, but, in principle, it could be profitably utilized. Our overall research mission is to understand CATs, to control CATs and eventually to engineer "smart" CATs. To this end, we combine tools from microscopy, molecular microbiolgy and systems and synthetic biology. We hope that our fundamental research findings will inspire novel biotechnical applications in probiotics research in the future.
Life cycle strategies: Spore quality-quantity tradeoff
We have developed high-end fluorescence timelapse microscopy assays and utilize novel fluorescent reporters which enable us to investigate the life cycle of sporulating bacteria with unprecedented detail. For example, our research on the B. subtilis life cycle has shown that sporulation and spore revival, which have been traditionally viewed as independent traits, are linked by a spore quality-quantity tradeoff. Under starvation stress B. subtilis bacteria can either make "more" or "better" spores. Similar tradeoffs are known to govern the production of progeny (seeds or eggs) in higher organisms. We are interested in the underlying molecular mechanisms that govern this tradeoff, its implications for the evolutionary adaptation of sporulating bacteria to different environments and how it affects the industrial production of spores for probiotics.
Read more here | Max-Planck-Gesellschaft (mpg.de)
Biophysical studies of cell-cell communication systems
In another line of research, we address the question how bacteria communicate and coordinate complex population behaviors with the help of diffusible signaling molecules and how this contributes to fitness. B. subtilis communicates by ComX-pheromones that activate the ComP/A two-component system and by multiple Phr-signaling peptides that are internalized to repress cytoplasmic Rap-receptors from the RRNPP-type family. We are investigating the function of these signaling networks by quantitative fluroescence microscopy, biochemical approaches and biophyiscal modeling. It is long known that bacteria can infer their population size with the help of chemical signals ("quorum sensing"). In a recent study we have utilized a novel FRET-based reporter and provided evidences that bacteria are capable to determine the composition of a mixed population by utilizing peptide-based pump-probe signaling systems that activate RRNPP-type receptors. These signaling systems are wide-spread in Gram-positive bacteria and could thus play an important role in regulating frequency dependent responses in heterogeneous populations.
Read more here | Max-Planck-Gesellschaft (mpg.de)
Cross- and interdisciplinary studies of stress-associated phenomena using B. subtilis as a model
Together with others we have recently investigated the hypothesis that reactive oxygen species could drive the formation of methane from methylated biomolecules. This route of methane formation could be common to all cells. We have tested central predictions of this theory using B. subtilis - spores and vegetative cells - as a model.
All organisms produce methane | Max-Planck-Gesellschaft (mpg.de)