Molecular Phytopathology
Prof. Dr. Regine Kahmann
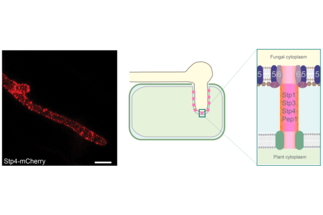
Regine Kahmann was director of the Department of Organismic Interactions between 2000 and 2019. Between April 2019 and June 2020 she served as acting head of the department and has since emeritus status. Her work was dedicated to the question how fungi colonize plants and cause disease. Her group has developed the biotrophic fungus Ustilago maydis that causes smut disease in corn (see Figure below) as a model to obtain molecular insights into how this fungus suppresses host immune responses and modulates plant processes to benefit the pathogen. Since the 2006 publication of the U. maydis genome sequence and detecting clustered genes encoding novel secreted protein effectors, her group has focused on the challenging functional analysis of such novel effectors and how a subset of them are taken up by cells of the host

Her most recent findings are listed below:
- We have discovered a surface-exposed protein complex of U. maydis that is essential for virulence and likely involved in effector delivery to host cells
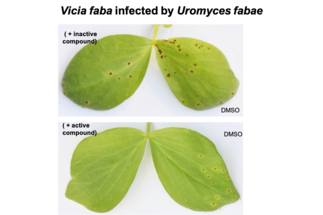
- We have developed the surface-exposed protein complex as new fungicide target
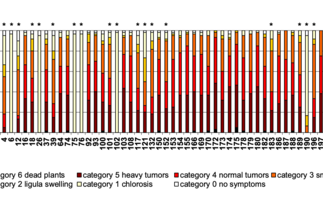
- We have used comparative genomics to identify core effectors with important virulence functions