Methylocystis sp. strain SC2
Our model organism, strain SC2, is able to grow on CH4 as sole source of carbon and energy. This type IIa methanotroph in the Alphaproteobacteria produces two isozymes of particulate methane monooxygenase (pMMO). These exhibit different methane oxidation kinetics and are encoded by the pmoCAB1 (low-affinity pMMO1) and pmoCAB2 (high-affinity pMMO2) operons (Baani and Liesack, 2008). The genome of strain SC2 was determined to comprise a 3.77 Mb circular chromosome and two large plasmids of 229.6 kb (pBSC2-1) and 143.5 kb (pBSC2-2) (Dam et al., 2012 and 2013).
Our previous transcriptome research allowed us to gain insights into differential gene expression in strain SC2 (Dam et al., 2014; Han et al., 2017). However, transcriptomics is time-consuming and does not allow for high-throughput analysis. Therefore, to be able to investigate the response of strain SC2 to experimental treatments on the molecular level more time-efficiently, we developed an analytical proteomics workflow for strain SC2. It tackles the major challenges related to the high amount of integral membrane proteins that need to be efficiently solubilized and digested for downstream analysis (Hakobyan et al., 2018). Each step of the workflow, including cell lysis, protein solubilization and digestion, and MS peptide quantification, was assessed and optimized. The workflow captures 62% (in total 2510 proteins) of the predicted SC2 proteome, with up to 10-fold increase in membrane-associated proteins relative to less effective conditions. Our new crude-lysate-MS approach proved not only to increase the proteome coverage of strain SC2, but also the protein quantification accuracy (global median CV of 3.2%). The newly developed proteomics workflow is now used to study the link between physiology and molecular biology of strain SC2.
The detection of cytosolic and membrane-bound hydrogenases in Methylocystis spp. prompted us to investigate whether strain SC2 is able to utilize hydrogen as an alternative energy source.
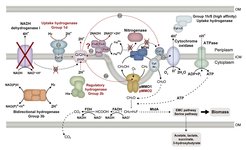
The addition of 2% H2 to the culture headspace had a significant effect on both growth yield and methane consumption rate under CH4 (6%) and O2 (3%) limited conditions (Hakobyan et al., 2020). The SC2 biomass yield doubled from 6.41 (±0.52) to 13.82 (±0.69) mg cell dry weight per mmol CH4, while total methane consumption significantly decreased. Complete hydrogen consumption was linked to treatment-specific changes in the SC2 proteome. In addition to hydrogenase accessory proteins, the production of Group 1d and Group 2b hydrogenases was significantly increased during the whole incubation period, in both short-term (19 h) and long-term (37 d) treatments. Transcript quantitation by real-time RT-PCR and SC2 proteomics showed that the Group 1d and Group 2b hydrogenases were coordinately regulated. Long-term hydrogen treatments (37 d) in a nitrogen-free environment induced a significant downregulation of NADH dehydrogenase I, in contrast to the constant upregulation of Group 1d hydrogenase. Therefore, we propose that during the long-term (37 d) incubation experiment, the functional role of complex I was taken over by Group 1d hydrogenase. Apparently, strain SC2 has the metabolic capacity to channel hydrogen-derived electrons into the quinone (Q/QH2) pool. Reduced quinone then transfers the electrons either to pMMO for methane oxidation or to cytochrome c and terminal oxidase for ATP generation (Figure).
In summary, the addition of 2% H2 to the culture headspace under CH4 and O2 limited conditions doubled the biomass yield of strain SC2, with less CH4 consumed. Thus, the utilization of hydrogen as an alternative energy source may be an effective strategy of Methylocystis spp. to increase their biomass yield and to survive under methane-limiting conditions. In fact, a strong negative correlation between low-affinity H2 oxidation and high-affinity CH4 oxidation at the soil-nodule interface led to the conclusion that in farmland and poplar soils, these two processes be performed by the same microbes. Given that in our study, strain SC2 oxidized hydrogen from the starting concentration (2%) to below the detection limit of the H2 sensor (0.02%) in batch cultivation mode, hydrogen-based mixotrophy may be relevant not only in the proximity of legume nodules, but also in the oxic-anoxic interfaces of methanogenic environments where methanotrophs encounter greater variations in methane, hydrogen, and oxygen concentrations (Hakobyan and Liesack, 2020).
