Bacterial Epitranscriptomics
Read- Write- Erase:
Understanding the biological role of NAD-capped RNAs in E. coli
In biological environments, cellular processes are typically regulated by interactions of proteins with RNA. Over recent decades, researchers tried to understand the molecular characteristics of RNA-protein interactions and how the protein discriminates between different potential binding sites on these RNAs. In general, proteins associate preferentially with defined RNA sequence or structure motifs, or a combination thereof. Another possibility for a protein to interact with RNA is the specific recognition of an RNA modification.
To date, more than 160 chemical modifications are known to decorate RNA molecules in eukaryotes and prokaryotes and thereby altering their diverse coding, structural, catalytic, and regulatory functions. In eukaryotes, the 7-methylguanosine cap protects mRNA from degradation and influences maturation, localisation, and translation. For decades, it was believed that a fundamental characteristic of prokaryotic gene expression was the absence of a RNA 5’-cap.
Recently we discovered the ubiquitous redox coenzyme NAD to be attached to a specific set of regulatory RNAs in bacteria in a cap-like manner, and to modulate the functions of these RNAs. Biochemical studies revealed that analogous to a eukaryotic cap, the 5′-NAD modification stabilizes RNA against degradation in vitro. Moreover, we identified the enzyme NudC to specifically remove the cap, a processes referred to as “decapping”. The identification of NAD-RNAs in all kingdoms of life opened a new chapter in the biology of prokaryotic and eukaryotic RNA modifications.
The recent advances in the field of RNA modifications clearly show that the epitranscriptome and its modifying enzymes form a complex constellation that holds widely diverse and probably still unknown functions. One of the next major scientific challenges will be the identification of the biological significance of this new RNA modification. In particular, the identification of the NAD-cap, suggests new biological roles of NAD in the context of RNA-protein interactions that regulate cellular processes.
Objectives
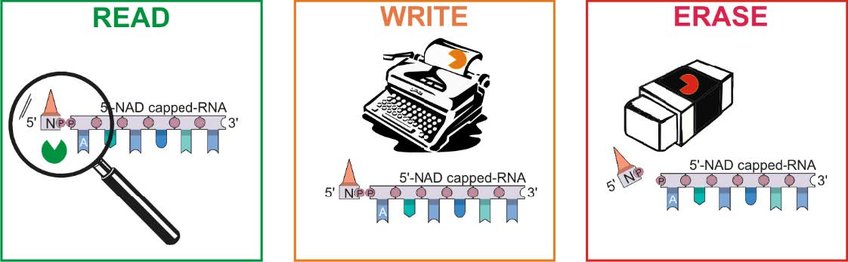
RNAs harbouring a 5‘-NAD-cap were found in all kingdoms of life, but except for an influence on RNA stability and processing not much is known about the biological consequences of this novel epitranscriptomic modification. We have the vision that the NAD-cap serves as a molecular hub for proteins.
Our research aims at discovering and characterizing new epitranscriptomic mechanisms of gene regulation based on NAD-RNAs in the model organism Escherichia coli. My inter-disciplinary, innovative goup focuses on the interaction of proteins with NAD-RNA to identify novel and important connections between redox biology, gene expression and regulation.
In particular, our research addresses the following questions:
I) Which proteins (readers, writers and erasers) specifically interact with NAD-capped-RNA?
II) Can NAD-RNA be post-transcriptionally modified by specific enzymes?
III) Which functional and physiological consequence have NAD-RNA/protein interaction in vitro and in vivo?
Considering the central role of the cofactor NAD in redox biochemistry, post-translational protein modification, and signalling, its attachment to RNA points to unknown roles of RNA in these processes and to yet undiscovered pathways in RNA metabolism and regulation. The identification of NAD as a new component of RNA might implement a new ‘epitranscriptomic’ layer of cellular regulation in all kingdoms of life.