
Evolutionary Biochemistry
Our lab studies the evolution of protein complexes at the interface between experimental biochemistry and evolutionary biology. The majority of proteins associate into higher-order complexes. They range from the simple - containing only a few genetically identical subunits – to the baroquely elaborate – containing dozens of genetically different subunits in precise arrangements. How and why complexes evolve and change in evolutionary history is largely unknown, but directly relates to major questions in evolutionary biology: Does biological complexity arise through many incremental steps, or through rare but drastic jumps? And is it always driven by natural selection for some beneficial function, or does blind chance play a role in creating and maintaining this complexity? Our lab combines experimental biochemistry with phylogenetic and bioinformatics tools to answer these questions on the scale of individual complexes as well as entire families of proteins and their interactions. In particular, we use ancestral sequence reconstruction to resurrect ancient complexes that last existed hundreds of millions of years ago. We then experimentally study these ancient proteins using mass photometry, electron microscopy, X-ray crystallography and functional assays to unravel the interplay of history, chance, and natural selection in producing the complexity that exists today.
Subtopics
Ancestral sequence reconstruction

Proteins have evolutionary histories as full of change, drama, and turmoil as the organisms that contain them. But unlike entire organisms, proteins do not fossilize and so we do not have direct access to their evolutionary past. But their origin stories are still encoded in their sequences, in the differences and similiarties shared by the same protein in different organisms. We can use this information to bring the past back to life. Literally. Ancestral sequence reconstruction uses a sequence alignment, a phylogenetic tree, and a simple model of sequence evolution to back-calculate the sequnce of proteins that existed hundreds of millions, or even billions of years ago (read our review on why this is useful here). We can then synthesize coding sequences for these proteins, express and purify them and study their properties in the lab. In that way, we can literally recapitulate billions of years of biochemical evolution in our test tubes.
Evolution of biological carbon fixation

The Calvin cycle enzyme Rubisco catalyzes the vast majority of all CO2 fixation on our planet. Because of its central role in the global carbon cycle, evolutionary transitions in exactly how this enzyme catalyzes its reaction have had dramatic consequences for history of life on earth. One theme of its history is its problematic side reaction with oxygen, which produces a toxic side-product that has to be detoxified in a process that significantly lowers photosynthetic yields. Rubisco first developed this problematic ability to react with oxygen almost unimaginably long ago: in the Archaean age, long before there was any oxygen in our planet’s atmosphere. As oxygenic photosynthesis started to oxygenate our planet, Rubisco had to repeatedly improve its ability to discriminate between CO2 and oxygen. It is thought that this occurred through a series of structural adaptations - including recruiting new subunits and new dedicated chaperones. Our lab is reconstructing the history of these structural elaborations (read our paper on one of the earliest elaborations here) to understand if and how they affected Rubiscos catalytic capabilities.
Evolution of new interfaces

How easily do new protein-protein interactions evolve – are they created by rare mutations that are subsequently preserved by selection? Or are complexes so common because they are mutationally very accessible? We are studying this question in the evolution of homomeric protein complexes and in particular citrate synthases. These enzymes catalyze the first step of the citric acid cycle in nearly all organisms, but their quaternary structure can vary dramatically. We have for example discovered one case in which this enzyme evolved to form striking assemblies resembling a famous fractal (see image on the left and read the paper here). But are such trajectories common and accessible to proteins at most times? To answer these questions we are developing high-throughput methods to measure the assembly state of thousands of protein variants. Such a deep mutational scans will map how close to assembly proteins are in the mutational space surrounding their sequence, and whether certain types of assembly are easier to evolve than others.
Complexity ratchets and reversibility of evolutionary transitions
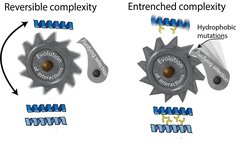
According to classical evolutionary theory, useless structures should quickly be lost to the constant onslaught of random, destructive mutations. Protein complexes may not follow this pattern: Their interfaces can become entrenched over time, making the protein reliant on assembly even if the interface does not directly contribute to function (read our review on this here). This results in a complexity ratchet: the evolution of a new interface is not easily reversible, because mutations that destroy the interface also diminish function (read our paper on a hydrophobic ratchet here). We study the mechanisms that produce such ratchets, their long-term evolutionary consequences, and how sometimes evolution overcomes such ratchets and reverts to simpler forms of assembly. We currently study the evolution of such ratchets in flagella and RubisCO.



