Function of the divisome
The divisome is a complex machinery composed of more than two dozen proteins. Some of its core components, such as FtsZ, are highly conserved among bacteria, whereas others are restricted to certain lineages, adapting divisome function to the specific physiological needs of the host. Despite decades of intensive research, the precise function of most core component and the mechanisms coordinating their activities are still unclear. Previous work on cell division has mainly focused on the model organism Escherichia coli. The essential divisome components identified in this organism are conserved in other bacteria, although their sequences are in some cases highly divergent. We have recently identified additional, new components of the C. crescentus divisome and revealed significant differences in the orchestration of the division process compared to the canonical E. coli system.
Investigating the machinery involved in cell wall constriction, we discovered in C. crescentus a previously uncharacterized regulatory protein that turned out to be a distant homolog of the known cell division protein FtsN. While FtsN was previously thought to be present only in E. coli and closely related species, we found that equivalent proteins are in fact present in all proteobacterial species (Möll & Thanbichler, 2009). Importantly, we were able to show that the cell wall-binding (SPOR) domain that is characteristic of FtsN, and widespread among many other proteins involved in cell wall biogenesis, acts as a general module mediating the recruitment of cell wall biosynthetic factors to the bacterial division site. Screening for interaction partners of C. crescentus FtsN, we have revealed a second factor with a key role in the coordination of the cell wall remodeling process, DipM (Möll et al., 2010). Interestingly, the function of DipM relies on a cell wall hydrolytic domain that has been rendered inactive by mutations in several catalytic residues. A similar situation has been seen in E. coli, where an unrelated protein bearing the same inactive hydrolase domain was shown to be part of a regulatory pathway coordinating cell wall constriction. However, this pathway is only conserved in close relatives of E. coli, indicating that the regulatory mechanisms at work in other bacteria must be different.
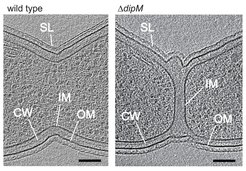
Function of DipM. Shown are virtual sections through electron cryo-tomograms of C. crescentus wild-type and dipM mutant cells. In the mutant, invagination of the inner and outer layers of the cell envelope are uncouples (IM: inner membrane, CW: cell wall, OM: outer membrane, SL: surface layer).
To elucidate the regulation of cell wall constriction in C. crescentus, we have recently initiated a comprehensive analysis of the numerous enzymes involved in peptidoglycan synthesis and degradation in this species. These studies have revealed that several additional peptidoglycan synthases and hydrolases of C. crescentus are recruited to the division site. However, in most cases, these proteins are not essential for the division process, because their loss can be compensated by various functionally equivalent proteins that are not specifically associated with the division apparatus (Strobel et al., 2014; Zielinska et al., 2017). Thus, the different growth modes of bacterial cells are mediated by a highly redundant set of enzymes with broad and overlapping functionality. We have now set out to determine how the activities of the different players are coordinated such as to ensure proper cell shape and cell wall integrity.
Apart from our work on peptidoglycan remodeling enzymes, we are currently studying other newly identified cell division proteins to clarify their biological function and their mode of interaction with other divisome components.
