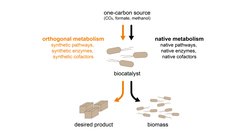
Synthetic cofactors and orthogonal metabolism
Synthetic cofactors
We define a synthetic cofactor as a compound similar in structure and chemical properties to a natural cofactor. A prominent example is nicotine mononucleotide (NMN), a derivate of NADH that already has been demonstrated to enable orthogonal metabolism. Our group aims to design additional synthetic analogues for cofactors involved in substrate selection and catalysis to enable a full separation of the catalyzed reaction from core metabolism. Here, we develop new synthesis routes to generate these cofactors and their derivates, and screen them in medium throughput against enzymes with affinity for the natural cofactor.
Synthetic enzymes
In order to adapt enzymes to synthetic cofactors, their active sites need to undergo large changes. Our approach to engineering enzymes with new cofactor selectivities is semi-rational, combining on our understanding of the active site with proposals made by computational models. We employ laboratory automation to screen variants in medium – to high throughput and work with the Metabolomics core facility for in-depth characterization of the best performing variants.
Synthetic pathways
We construct our pathways in a modular fashion, allowing a level of plug-and-play to combine different substrate valorization with production routes. This allows a step-wise implementation of our cascades in vivo. In addition, we engineer production routes for our synthetic cofactors so there is no need to supply them to the culture medium. Here, we combine our expertise in metabolic engineering with the analytics provided by the Proteomics core facility to fine-tune enzyme levels for optimal pathway performance. Our set goal is the adjustment of a production strain to a fully orthogonal cascade at scale.
