Bacterial development & differentiation
Prof. Dr. Lotte Søgaard-Andersen
Research area
The overall goal of our research is to unravel the mechanisms that allow bacteria to adapt and differentiate in response to changes in the environment. Bacteria have evolved at least three strategies that allow them to cope with such changes. One strategy centers on changes in gene expression ranging from changes in the expression of relatively few genes to changes in the expression of large numbers of genes culminating in cell differentiation. A second strategy centers on changes in the motility behavior of cells. Finally, a third strategy centers on changes to the cell cycle. To implement these strategies bacterial cells have to process vast amounts of information and then generate the appropriate output response. Information processing is carried out by complex networks of signal transduction proteins. A challenging problem in bacterial adaptation and differentiation is to understand how these protein networks are organized in space and time to allow the ordered execution of various tasks.
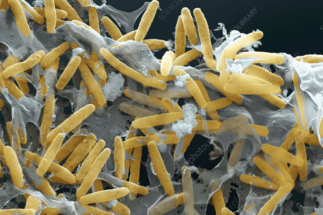
We use Myxococcus xanthus as a wonderful model system to study bacterial adaptation and differentiation. In particular, we study the signal transduction pathways and networks governing differentiation, motility and the cell cycle. In parallel approaches, we aim to understand how molecular machineries involved in motility and cell division function.