Bacterial Secretion Systems
Dr. Andreas Diepold
Research Area

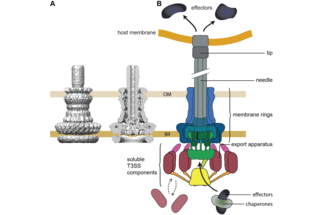
Center, schematic depiction of the type III secretion system. Main substructures indicated; HM, host membrane; OM, outer membrane; IM, inner membrane. Current research foci of our group are depicted on the left and right side, arranged from projects investigating the molecular understanding of the T3SS (bottom, red font) to projects focused on its physiological role during infection (top, blue font).
To interact with their environment, bacteria use can use secretion systems. These systems are essential for many aspects of bacterial communication, symbiosis, competition, and virulence. Bacteria that live in contact with eukaryotic cells can manipulate the behavior of these target cells using the type III secretion system (T3SS), a molecular syringe also known as "injectisome", which injects effector proteins into host cells.
The T3SS is essential for virulence in many important human pathogens, including Salmonella, Shigella, and pathogenic Escherichia coli, that cause several millions of deaths per year. It is also important in hospital infections, such as by Pseudomonas aeruginosa, where presence of a functional T3SS is associated with higher mortality in animal models and increased antibiotic resistance, severe disease, and a bad prognosis in infected humans.
While the translocated effector proteins vary between different bacterial species, the T3SS itself is highly conserved, making it an attractive target for anti-virulence therapeutics. However, although parts of the evocative "injection device" structure of the injectisome have been characterized in great detail, we know surprisingly little about the molecular function of the T3SS. How do the components of the injectisome interact to allow the fast and ordered translocation of the effectors? Which molecular events lead to effector export? And how does the injectisome respond to external signals?
My group wants to understand how the dynamic T3SS works on the molecular level, how it is activated and regulated during the infection process, how function and regulation of the T3SS contribute to virulence and how virulence factors are co-regulated and influence each other during infection.
We aim to uncover molecular effects and correlations and link these to physiological impact, such as translocation rate into host cells. Accordingly, we apply a wide range of methods, both in our lab and in collaborations with leading researchers within and beyond the Max Planck Institute. These range from cutting-edge live-cell and superresolution microscopy, complemented by various biochemical and genetic methods, to cell culture-based infection assays. Wherever possible, we analyze the T3SS in live bacteria, both under controlled conditions and in contact to host cells. This approach allows us to directly test for the physiological significance of our findings for T3SS function and successful infection.